#PCA is your friend, part two
Continued from the post below
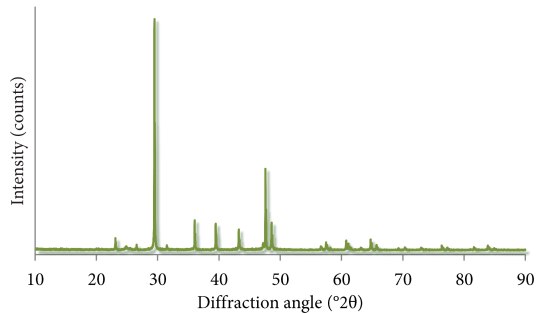
The loadings the thing, wherin I’ll catch the essence of the dataset. Principal component analysis (PCA) is an excellent, and often essential, method for analysing a large amount of data. Our research question centers around the differences between two fossil sites, and the large dataset we have at hand is made up of x-ray fluorescence spectra from fossil bones. These data go into the PCA, and out pour our beautiful results in two forms – scores and loadings.
Let’s look at the loadings.
Loadings let us see the major sources of variation in our dataset. By ‘sources of variation’, here I mean the way in which spectra differ from one another, like having peaks in different places. Different sources of variation are teased out for each principal component, and we can visualise these ‘components’ with the loadings. Take the loadings for PC1, for example:
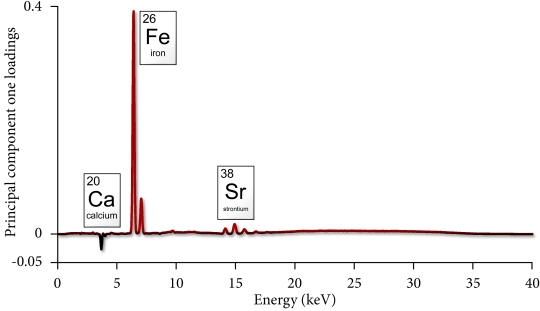
Principal component one loadings for x-ray fluorescence spectra. Data were collected from fossil antelope bone. Modified from Thomas and Chinsamy 2011.
These loadings show us that the peaks attributed to iron and strontium are positively weighted, and the peaks attributed to calcium are negatively weighted. So this means that some samples in our dataset that have a great deal of Fe and Sr, and some samples have extra calcium, over and above the amount typical amount for bone. Now we jump to the next step, and used our PC1 loadings to interpret our PC1 scores, which are below:
We found that our Fe and Sr peaks were positively loaded. In our scores plot, this means that samples with positive PC1 scores should be rich with Fe and Sr. Likewise, our Ca peaks were negatively weighted, and so our samples with negative PC1 scores probably contain an extra calcite mineral. If we take a look at our PC1 scores we find that the positively weighted samples are all from Elandsfontein Main, and all of the Swartklip 1 have negative score values.
So we have found a chemical difference between the bones of these two sites. Elandsfontein bones have been infiltrated with iron and strontium rich minerals, which actually turn out to be clays and sands deposited by groundwater. The Swartklip bones contain abundant calcite. What does that mean for the burial history of these two sites?
The fossil bones at Elandsfontein Main and Swartklip 1 both accumulated in dune environments during the Pleistocene. The Elandsfontein Main site remained inland, and the slightly acidic groundwater that percolated through the fossils partially dissolved the bones and filled them with sediment. In contrast, sea level change periodically brought the coastline close to the Swartklip 1 site, where it is now, actually. The marine influence introduced calcium carbonate into the environment, which buffered the acidic groundwater and laced it with dissolved carbonates. These carbonates precipitated onto the Swartklip 1 fossils.
So, at Elandsfontein Main we have fossils that have been subjected to acidic groundwater for tens of thousands of years, and at Swartklip 1, we have fossils that have been periodically buffered by soil carbonates. If I was to pick one site to start looking for intact and well preserved bone, even down to isotope-level, I would start with the fossils Swartklip 1.
So yeah, this is the type of information we get from spectroscopy and principal components analysis. Pretty cool eh.
Thomas D B, Chinsamy A, 2011. Chemometric analysis of EDXRF measurements from fossil bone. X-ray Spectrometry 40: 441-445
Still to come: R code for pretreating spectra, performing a PCA analysis, and producing informative graphs…